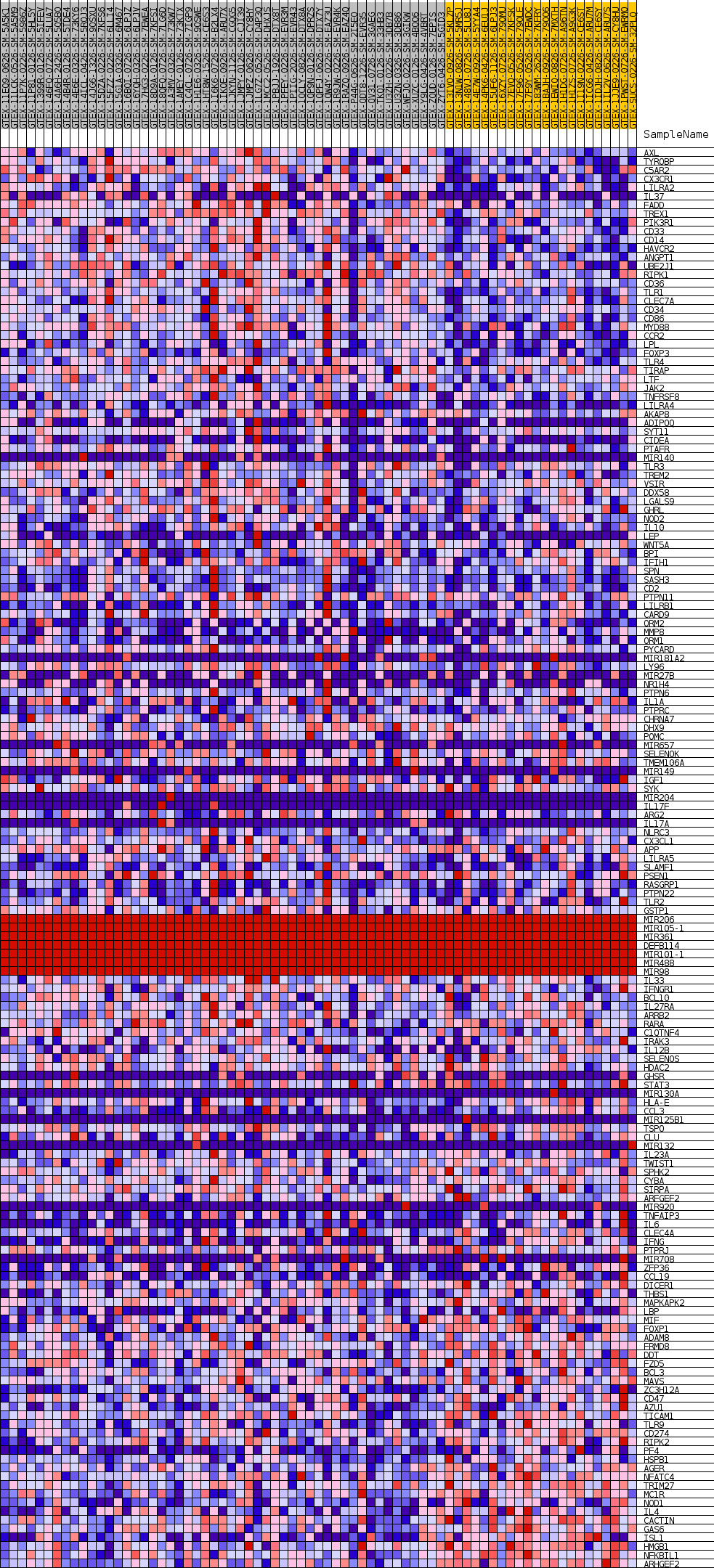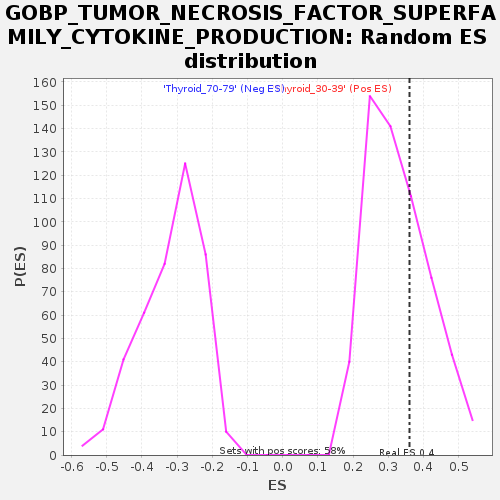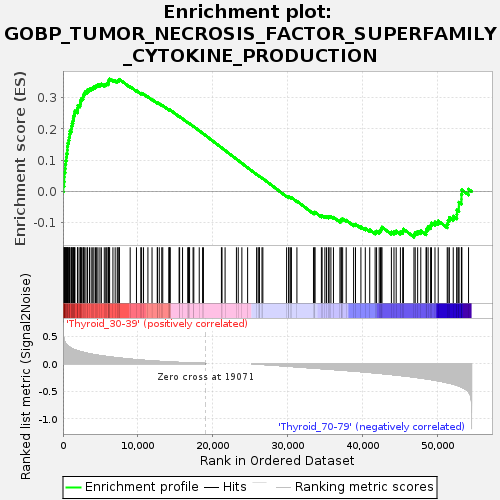
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_TUMOR_NECROSIS_FACTOR_SUPERFAMILY_CYTOKINE_PRODUCTION |
| Enrichment Score (ES) | 0.36038846 |
| Normalized Enrichment Score (NES) | 1.1011841 |
| Nominal p-value | 0.32413793 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AXL | NA | 53 | 0.499 | 0.0173 | Yes |
| 2 | TYROBP | NA | 152 | 0.425 | 0.0310 | Yes |
| 3 | C5AR2 | NA | 167 | 0.419 | 0.0461 | Yes |
| 4 | CX3CR1 | NA | 174 | 0.417 | 0.0613 | Yes |
| 5 | LILRA2 | NA | 281 | 0.382 | 0.0733 | Yes |
| 6 | IL37 | NA | 309 | 0.374 | 0.0865 | Yes |
| 7 | FADD | NA | 376 | 0.360 | 0.0984 | Yes |
| 8 | TREX1 | NA | 437 | 0.350 | 0.1101 | Yes |
| 9 | PIK3R1 | NA | 488 | 0.341 | 0.1217 | Yes |
| 10 | CD33 | NA | 578 | 0.327 | 0.1320 | Yes |
| 11 | CD14 | NA | 585 | 0.326 | 0.1439 | Yes |
| 12 | HAVCR2 | NA | 641 | 0.321 | 0.1546 | Yes |
| 13 | ANGPT1 | NA | 756 | 0.309 | 0.1638 | Yes |
| 14 | UBE2J1 | NA | 819 | 0.304 | 0.1738 | Yes |
| 15 | RIPK1 | NA | 880 | 0.297 | 0.1836 | Yes |
| 16 | CD36 | NA | 926 | 0.292 | 0.1934 | Yes |
| 17 | TLR1 | NA | 1104 | 0.276 | 0.2003 | Yes |
| 18 | CLEC7A | NA | 1154 | 0.273 | 0.2094 | Yes |
| 19 | CD34 | NA | 1216 | 0.269 | 0.2181 | Yes |
| 20 | CD86 | NA | 1325 | 0.261 | 0.2257 | Yes |
| 21 | MYD88 | NA | 1367 | 0.259 | 0.2344 | Yes |
| 22 | CCR2 | NA | 1430 | 0.255 | 0.2426 | Yes |
| 23 | LPL | NA | 1537 | 0.249 | 0.2498 | Yes |
| 24 | FOXP3 | NA | 1596 | 0.246 | 0.2577 | Yes |
| 25 | TLR4 | NA | 1930 | 0.231 | 0.2600 | Yes |
| 26 | TIRAP | NA | 1967 | 0.229 | 0.2677 | Yes |
| 27 | LTF | NA | 1983 | 0.229 | 0.2758 | Yes |
| 28 | JAK2 | NA | 2264 | 0.218 | 0.2787 | Yes |
| 29 | TNFRSF8 | NA | 2317 | 0.216 | 0.2856 | Yes |
| 30 | LILRA4 | NA | 2371 | 0.214 | 0.2925 | Yes |
| 31 | AKAP8 | NA | 2482 | 0.210 | 0.2981 | Yes |
| 32 | ADIPOQ | NA | 2669 | 0.203 | 0.3022 | Yes |
| 33 | SYT11 | NA | 2726 | 0.201 | 0.3085 | Yes |
| 34 | CIDEA | NA | 2826 | 0.198 | 0.3139 | Yes |
| 35 | PTAFR | NA | 2949 | 0.194 | 0.3188 | Yes |
| 36 | MIR140 | NA | 3211 | 0.186 | 0.3208 | Yes |
| 37 | TLR3 | NA | 3286 | 0.184 | 0.3261 | Yes |
| 38 | TREM2 | NA | 3542 | 0.176 | 0.3279 | Yes |
| 39 | VSIR | NA | 3766 | 0.170 | 0.3300 | Yes |
| 40 | DDX58 | NA | 3977 | 0.165 | 0.3322 | Yes |
| 41 | LGALS9 | NA | 4133 | 0.161 | 0.3353 | Yes |
| 42 | GHRL | NA | 4348 | 0.156 | 0.3370 | Yes |
| 43 | NOD2 | NA | 4498 | 0.152 | 0.3399 | Yes |
| 44 | IL10 | NA | 4670 | 0.149 | 0.3422 | Yes |
| 45 | LEP | NA | 4936 | 0.143 | 0.3425 | Yes |
| 46 | WNT5A | NA | 5125 | 0.140 | 0.3442 | Yes |
| 47 | BPI | NA | 5515 | 0.132 | 0.3419 | Yes |
| 48 | IFIH1 | NA | 5715 | 0.128 | 0.3429 | Yes |
| 49 | SPN | NA | 5840 | 0.126 | 0.3452 | Yes |
| 50 | SASH3 | NA | 6046 | 0.122 | 0.3460 | Yes |
| 51 | CD2 | NA | 6053 | 0.122 | 0.3503 | Yes |
| 52 | PTPN11 | NA | 6065 | 0.122 | 0.3546 | Yes |
| 53 | LILRB1 | NA | 6157 | 0.121 | 0.3573 | Yes |
| 54 | CARD9 | NA | 6229 | 0.119 | 0.3604 | Yes |
| 55 | ORM2 | NA | 6671 | 0.112 | 0.3564 | No |
| 56 | MMP8 | NA | 6969 | 0.107 | 0.3548 | No |
| 57 | ORM1 | NA | 7246 | 0.103 | 0.3535 | No |
| 58 | PYCARD | NA | 7352 | 0.101 | 0.3553 | No |
| 59 | MIR181A2 | NA | 7488 | 0.099 | 0.3565 | No |
| 60 | LY96 | NA | 7534 | 0.098 | 0.3592 | No |
| 61 | MIR27B | NA | 8975 | 0.078 | 0.3356 | No |
| 62 | NR1H4 | NA | 9816 | 0.068 | 0.3227 | No |
| 63 | PTPN6 | NA | 10393 | 0.062 | 0.3144 | No |
| 64 | IL1A | NA | 10503 | 0.060 | 0.3146 | No |
| 65 | PTPRC | NA | 10760 | 0.058 | 0.3120 | No |
| 66 | CHRNA7 | NA | 11346 | 0.052 | 0.3031 | No |
| 67 | DHX9 | NA | 11895 | 0.046 | 0.2947 | No |
| 68 | POMC | NA | 12604 | 0.040 | 0.2832 | No |
| 69 | MIR657 | NA | 12613 | 0.040 | 0.2845 | No |
| 70 | SELENOK | NA | 12864 | 0.038 | 0.2813 | No |
| 71 | TMEM106A | NA | 13195 | 0.035 | 0.2765 | No |
| 72 | MIR149 | NA | 13329 | 0.034 | 0.2753 | No |
| 73 | IGF1 | NA | 14153 | 0.028 | 0.2612 | No |
| 74 | SYK | NA | 14202 | 0.028 | 0.2613 | No |
| 75 | MIR204 | NA | 14204 | 0.028 | 0.2623 | No |
| 76 | IL17F | NA | 14335 | 0.027 | 0.2609 | No |
| 77 | ARG2 | NA | 15513 | 0.019 | 0.2400 | No |
| 78 | IL17A | NA | 15583 | 0.018 | 0.2394 | No |
| 79 | NLRC3 | NA | 15973 | 0.016 | 0.2328 | No |
| 80 | CX3CL1 | NA | 16665 | 0.012 | 0.2206 | No |
| 81 | APP | NA | 16779 | 0.012 | 0.2189 | No |
| 82 | LILRA5 | NA | 16820 | 0.012 | 0.2186 | No |
| 83 | SLAMF1 | NA | 16912 | 0.011 | 0.2174 | No |
| 84 | PSEN1 | NA | 17379 | 0.009 | 0.2091 | No |
| 85 | RASGRP1 | NA | 17478 | 0.009 | 0.2076 | No |
| 86 | PTPN22 | NA | 18203 | 0.005 | 0.1945 | No |
| 87 | TLR2 | NA | 18649 | 0.003 | 0.1865 | No |
| 88 | GSTP1 | NA | 18764 | 0.003 | 0.1845 | No |
| 89 | MIR206 | NA | 21167 | 0.000 | 0.1404 | No |
| 90 | MIR105-1 | NA | 21246 | 0.000 | 0.1389 | No |
| 91 | MIR361 | NA | 21657 | 0.000 | 0.1314 | No |
| 92 | DEFB114 | NA | 23172 | 0.000 | 0.1036 | No |
| 93 | MIR101-1 | NA | 23415 | 0.000 | 0.0991 | No |
| 94 | MIR488 | NA | 23899 | 0.000 | 0.0903 | No |
| 95 | MIR98 | NA | 24667 | 0.000 | 0.0762 | No |
| 96 | IL33 | NA | 25866 | -0.007 | 0.0544 | No |
| 97 | IFNGR1 | NA | 26069 | -0.010 | 0.0511 | No |
| 98 | BCL10 | NA | 26199 | -0.011 | 0.0491 | No |
| 99 | IL27RA | NA | 26246 | -0.011 | 0.0487 | No |
| 100 | ARRB2 | NA | 26587 | -0.014 | 0.0429 | No |
| 101 | RARA | NA | 26705 | -0.016 | 0.0414 | No |
| 102 | C1QTNF4 | NA | 29876 | -0.046 | -0.0152 | No |
| 103 | IRAK3 | NA | 30155 | -0.048 | -0.0186 | No |
| 104 | IL12B | NA | 30171 | -0.049 | -0.0171 | No |
| 105 | SELENOS | NA | 30376 | -0.051 | -0.0189 | No |
| 106 | HDAC2 | NA | 30522 | -0.052 | -0.0197 | No |
| 107 | GHSR | NA | 31254 | -0.059 | -0.0310 | No |
| 108 | STAT3 | NA | 33467 | -0.081 | -0.0687 | No |
| 109 | MIR130A | NA | 33636 | -0.082 | -0.0687 | No |
| 110 | HLA-E | NA | 33665 | -0.083 | -0.0662 | No |
| 111 | CCL3 | NA | 34524 | -0.091 | -0.0786 | No |
| 112 | MIR125B1 | NA | 34621 | -0.092 | -0.0770 | No |
| 113 | TSPO | NA | 34989 | -0.096 | -0.0802 | No |
| 114 | CLU | NA | 35202 | -0.098 | -0.0805 | No |
| 115 | MIR132 | NA | 35454 | -0.100 | -0.0815 | No |
| 116 | IL23A | NA | 35556 | -0.102 | -0.0796 | No |
| 117 | TWIST1 | NA | 35796 | -0.104 | -0.0802 | No |
| 118 | SPHK2 | NA | 36137 | -0.107 | -0.0825 | No |
| 119 | CYBA | NA | 37003 | -0.116 | -0.0942 | No |
| 120 | SIRPA | NA | 37171 | -0.117 | -0.0930 | No |
| 121 | ARFGEF2 | NA | 37173 | -0.117 | -0.0887 | No |
| 122 | MIR920 | NA | 37364 | -0.120 | -0.0878 | No |
| 123 | TNFAIP3 | NA | 37838 | -0.124 | -0.0919 | No |
| 124 | IL6 | NA | 38829 | -0.135 | -0.1052 | No |
| 125 | CLEC4A | NA | 39082 | -0.138 | -0.1048 | No |
| 126 | IFNG | NA | 39805 | -0.146 | -0.1127 | No |
| 127 | PTPRJ | NA | 40391 | -0.153 | -0.1178 | No |
| 128 | MIR708 | NA | 40983 | -0.160 | -0.1228 | No |
| 129 | ZFP36 | NA | 41704 | -0.169 | -0.1298 | No |
| 130 | CCL19 | NA | 41896 | -0.172 | -0.1271 | No |
| 131 | DICER1 | NA | 42249 | -0.176 | -0.1271 | No |
| 132 | THBS1 | NA | 42420 | -0.179 | -0.1237 | No |
| 133 | MAPKAPK2 | NA | 42521 | -0.180 | -0.1189 | No |
| 134 | LBP | NA | 42622 | -0.181 | -0.1141 | No |
| 135 | MIF | NA | 43879 | -0.198 | -0.1299 | No |
| 136 | FOXP1 | NA | 44223 | -0.203 | -0.1288 | No |
| 137 | ADAM8 | NA | 44518 | -0.207 | -0.1266 | No |
| 138 | FRMD8 | NA | 45081 | -0.215 | -0.1291 | No |
| 139 | DDT | NA | 45389 | -0.220 | -0.1267 | No |
| 140 | FZD5 | NA | 45487 | -0.222 | -0.1203 | No |
| 141 | BCL3 | NA | 46892 | -0.245 | -0.1371 | No |
| 142 | MAVS | NA | 47058 | -0.249 | -0.1311 | No |
| 143 | ZC3H12A | NA | 47386 | -0.254 | -0.1278 | No |
| 144 | CD47 | NA | 47805 | -0.262 | -0.1259 | No |
| 145 | AZU1 | NA | 48507 | -0.276 | -0.1287 | No |
| 146 | TICAM1 | NA | 48576 | -0.277 | -0.1198 | No |
| 147 | TLR9 | NA | 48781 | -0.282 | -0.1133 | No |
| 148 | CD274 | NA | 49116 | -0.289 | -0.1088 | No |
| 149 | RIPK2 | NA | 49236 | -0.292 | -0.1003 | No |
| 150 | PF4 | NA | 49702 | -0.302 | -0.0978 | No |
| 151 | HSPB1 | NA | 50134 | -0.312 | -0.0943 | No |
| 152 | AGER | NA | 51337 | -0.346 | -0.1037 | No |
| 153 | NFATC4 | NA | 51446 | -0.350 | -0.0929 | No |
| 154 | TRIM27 | NA | 51623 | -0.355 | -0.0831 | No |
| 155 | MC1R | NA | 52142 | -0.374 | -0.0790 | No |
| 156 | NOD1 | NA | 52624 | -0.392 | -0.0735 | No |
| 157 | IL4 | NA | 52635 | -0.393 | -0.0593 | No |
| 158 | CACTIN | NA | 52893 | -0.405 | -0.0492 | No |
| 159 | GAS6 | NA | 52894 | -0.405 | -0.0344 | No |
| 160 | ISL1 | NA | 53237 | -0.426 | -0.0251 | No |
| 161 | HMGB1 | NA | 53239 | -0.426 | -0.0095 | No |
| 162 | NFKBIL1 | NA | 53297 | -0.429 | 0.0051 | No |
| 163 | ARHGEF2 | NA | 54186 | -0.510 | 0.0074 | No |