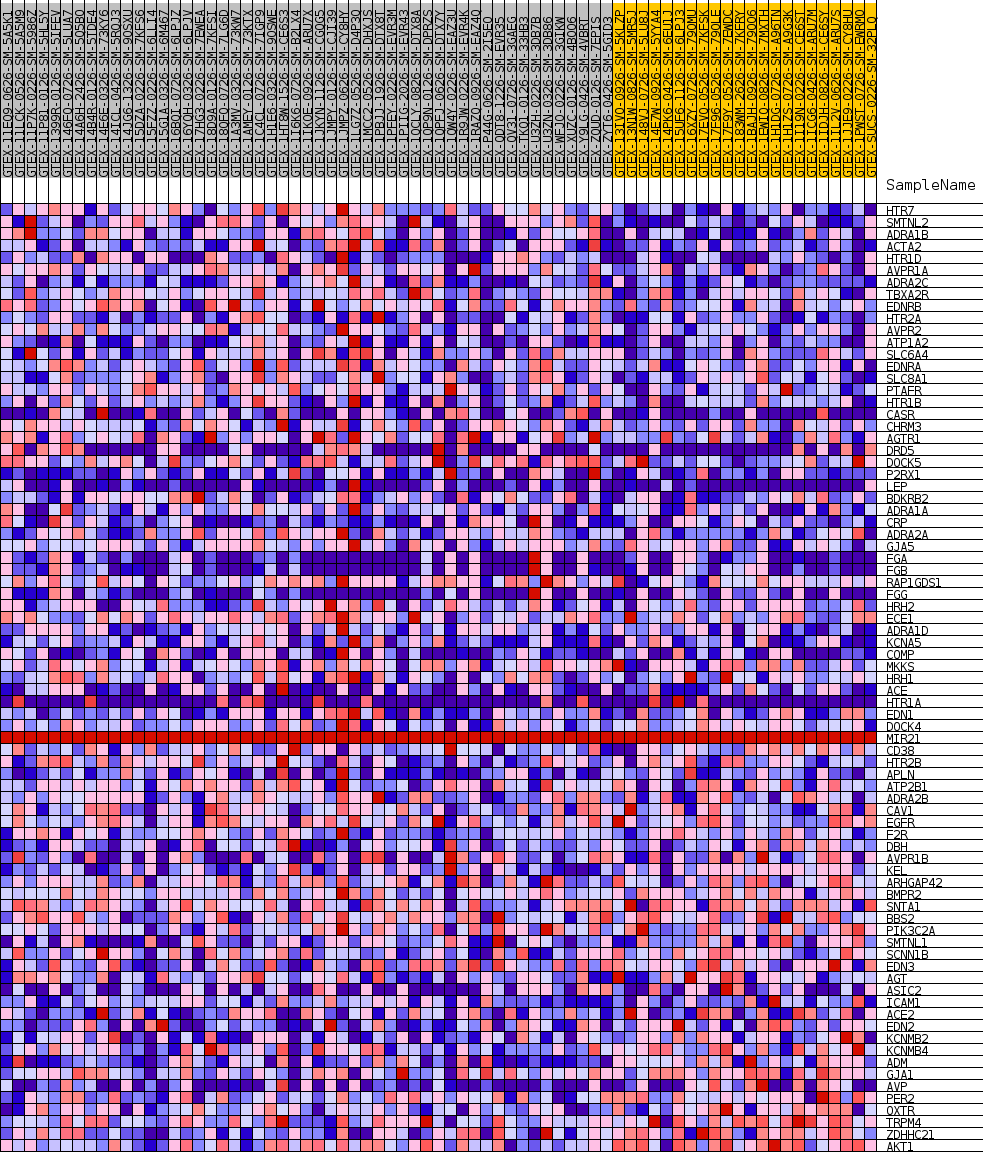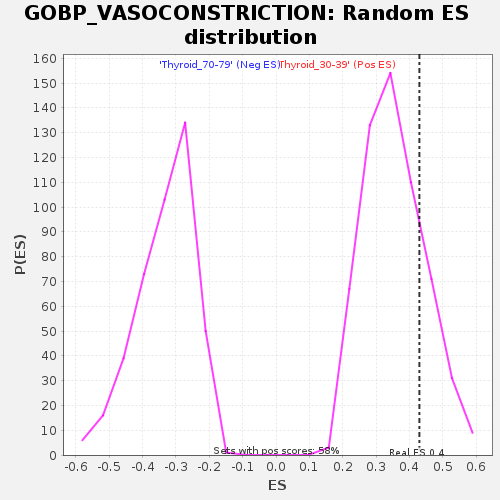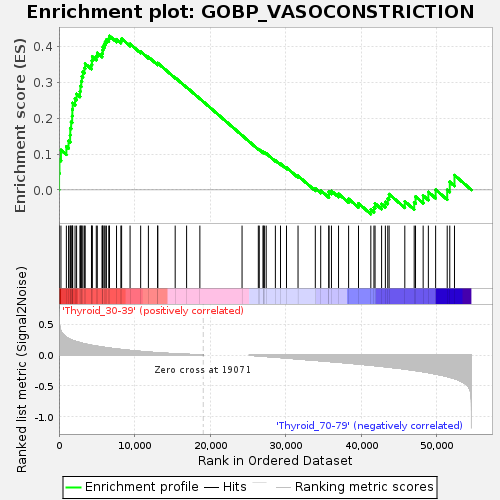
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_VASOCONSTRICTION |
| Enrichment Score (ES) | 0.42966735 |
| Normalized Enrichment Score (NES) | 1.2158208 |
| Nominal p-value | 0.20415226 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HTR7 | NA | 8 | 0.587 | 0.0463 | Yes |
| 2 | SMTNL2 | NA | 60 | 0.491 | 0.0842 | Yes |
| 3 | ADRA1B | NA | 257 | 0.388 | 0.1113 | Yes |
| 4 | ACTA2 | NA | 975 | 0.288 | 0.1209 | Yes |
| 5 | HTR1D | NA | 1264 | 0.265 | 0.1366 | Yes |
| 6 | AVPR1A | NA | 1472 | 0.253 | 0.1528 | Yes |
| 7 | ADRA2C | NA | 1490 | 0.252 | 0.1724 | Yes |
| 8 | TBXA2R | NA | 1607 | 0.246 | 0.1898 | Yes |
| 9 | EDNRB | NA | 1742 | 0.239 | 0.2062 | Yes |
| 10 | HTR2A | NA | 1767 | 0.238 | 0.2245 | Yes |
| 11 | AVPR2 | NA | 1812 | 0.236 | 0.2424 | Yes |
| 12 | ATP1A2 | NA | 2125 | 0.224 | 0.2544 | Yes |
| 13 | SLC6A4 | NA | 2329 | 0.215 | 0.2677 | Yes |
| 14 | EDNRA | NA | 2777 | 0.200 | 0.2753 | Yes |
| 15 | SLC8A1 | NA | 2834 | 0.198 | 0.2899 | Yes |
| 16 | PTAFR | NA | 2949 | 0.194 | 0.3032 | Yes |
| 17 | HTR1B | NA | 3027 | 0.191 | 0.3169 | Yes |
| 18 | CASR | NA | 3163 | 0.187 | 0.3292 | Yes |
| 19 | CHRM3 | NA | 3359 | 0.182 | 0.3400 | Yes |
| 20 | AGTR1 | NA | 3466 | 0.178 | 0.3522 | Yes |
| 21 | DRD5 | NA | 4299 | 0.157 | 0.3494 | Yes |
| 22 | DOCK5 | NA | 4359 | 0.156 | 0.3606 | Yes |
| 23 | P2RX1 | NA | 4404 | 0.155 | 0.3720 | Yes |
| 24 | LEP | NA | 4936 | 0.143 | 0.3736 | Yes |
| 25 | BDKRB2 | NA | 5068 | 0.141 | 0.3824 | Yes |
| 26 | ADRA1A | NA | 5698 | 0.129 | 0.3810 | Yes |
| 27 | CRP | NA | 5763 | 0.127 | 0.3899 | Yes |
| 28 | ADRA2A | NA | 5817 | 0.126 | 0.3989 | Yes |
| 29 | GJA5 | NA | 5982 | 0.124 | 0.4057 | Yes |
| 30 | FGA | NA | 6108 | 0.121 | 0.4130 | Yes |
| 31 | FGB | NA | 6275 | 0.118 | 0.4193 | Yes |
| 32 | RAP1GDS1 | NA | 6590 | 0.113 | 0.4225 | Yes |
| 33 | FGG | NA | 6684 | 0.112 | 0.4297 | Yes |
| 34 | HRH2 | NA | 7615 | 0.097 | 0.4203 | No |
| 35 | ECE1 | NA | 8193 | 0.089 | 0.4168 | No |
| 36 | ADRA1D | NA | 8289 | 0.088 | 0.4220 | No |
| 37 | KCNA5 | NA | 9409 | 0.073 | 0.4072 | No |
| 38 | COMP | NA | 10810 | 0.057 | 0.3861 | No |
| 39 | MKKS | NA | 11834 | 0.047 | 0.3710 | No |
| 40 | HRH1 | NA | 13051 | 0.036 | 0.3516 | No |
| 41 | ACE | NA | 13079 | 0.036 | 0.3539 | No |
| 42 | HTR1A | NA | 15380 | 0.020 | 0.3133 | No |
| 43 | EDN1 | NA | 16892 | 0.011 | 0.2865 | No |
| 44 | DOCK4 | NA | 18648 | 0.003 | 0.2545 | No |
| 45 | MIR21 | NA | 24232 | 0.000 | 0.1521 | No |
| 46 | CD38 | NA | 26386 | -0.013 | 0.1136 | No |
| 47 | HTR2B | NA | 26510 | -0.014 | 0.1124 | No |
| 48 | APLN | NA | 27026 | -0.018 | 0.1045 | No |
| 49 | ATP2B1 | NA | 27034 | -0.018 | 0.1058 | No |
| 50 | ADRA2B | NA | 27188 | -0.020 | 0.1046 | No |
| 51 | CAV1 | NA | 27428 | -0.022 | 0.1019 | No |
| 52 | EGFR | NA | 28643 | -0.033 | 0.0823 | No |
| 53 | F2R | NA | 29322 | -0.040 | 0.0730 | No |
| 54 | DBH | NA | 30115 | -0.048 | 0.0623 | No |
| 55 | AVPR1B | NA | 31643 | -0.063 | 0.0393 | No |
| 56 | KEL | NA | 33930 | -0.085 | 0.0041 | No |
| 57 | ARHGAP42 | NA | 34644 | -0.093 | -0.0016 | No |
| 58 | BMPR2 | NA | 35722 | -0.103 | -0.0132 | No |
| 59 | SNTA1 | NA | 35730 | -0.103 | -0.0052 | No |
| 60 | BBS2 | NA | 36068 | -0.107 | -0.0029 | No |
| 61 | PIK3C2A | NA | 37005 | -0.116 | -0.0109 | No |
| 62 | SMTNL1 | NA | 38338 | -0.130 | -0.0251 | No |
| 63 | SCNN1B | NA | 39647 | -0.144 | -0.0376 | No |
| 64 | EDN3 | NA | 41280 | -0.164 | -0.0546 | No |
| 65 | AGT | NA | 41692 | -0.169 | -0.0488 | No |
| 66 | ASIC2 | NA | 41839 | -0.171 | -0.0379 | No |
| 67 | ICAM1 | NA | 42705 | -0.182 | -0.0394 | No |
| 68 | ACE2 | NA | 43205 | -0.188 | -0.0336 | No |
| 69 | EDN2 | NA | 43523 | -0.193 | -0.0242 | No |
| 70 | KCNMB2 | NA | 43701 | -0.196 | -0.0119 | No |
| 71 | KCNMB4 | NA | 45778 | -0.226 | -0.0321 | No |
| 72 | ADM | NA | 47020 | -0.248 | -0.0352 | No |
| 73 | GJA1 | NA | 47196 | -0.251 | -0.0186 | No |
| 74 | AVP | NA | 48206 | -0.270 | -0.0158 | No |
| 75 | PER2 | NA | 48895 | -0.284 | -0.0059 | No |
| 76 | OXTR | NA | 49858 | -0.306 | 0.0007 | No |
| 77 | TRPM4 | NA | 51390 | -0.348 | 0.0001 | No |
| 78 | ZDHHC21 | NA | 51731 | -0.359 | 0.0223 | No |
| 79 | AKT1 | NA | 52354 | -0.381 | 0.0410 | No |