
|
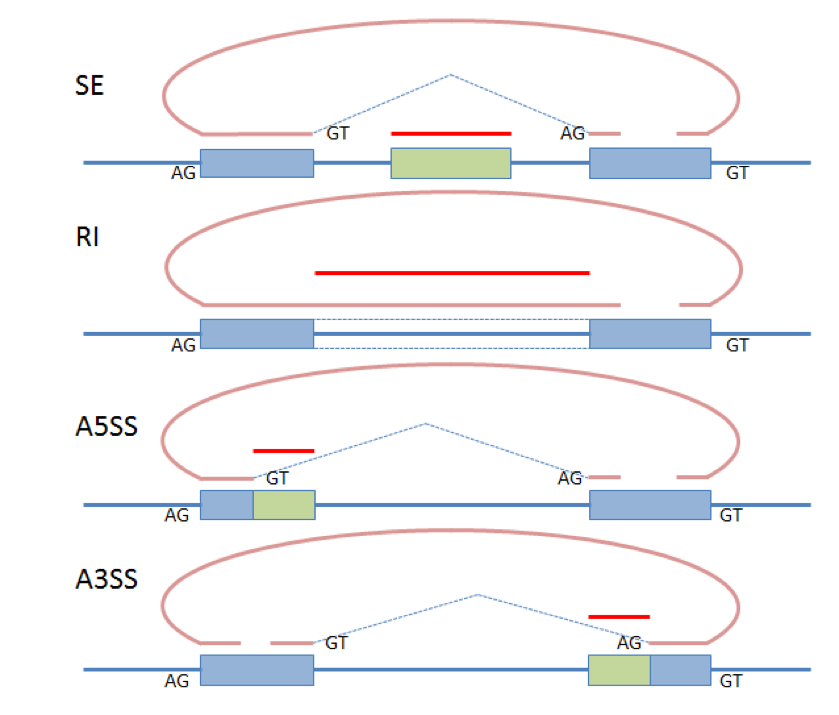
CircSplice is a custom script which developed a de novo algorithm for circRNA alternative splicing (circ-AS) detection across samples. CircSplice considers 4 types: skipping exons (SE), retained intron (RI), alternative 5' splice site (A5SS) and alternative 3' splice site (A3SS) in back-splicing read and corresponding paired read. CircSplice also considers splice sites GT-AG or CT-AC. Innovatively, CircSplice supports comparing circRNA AS events across different samples. For example, user can compare circRNA AS from cancer and adjacent normal tissues and detect the cancer-specific circRNA AS. CircSplice is implemented in Perl and has been tested in Unix and Mac OS X. |
 |